- Details
-
Thursday, 19 March 2015
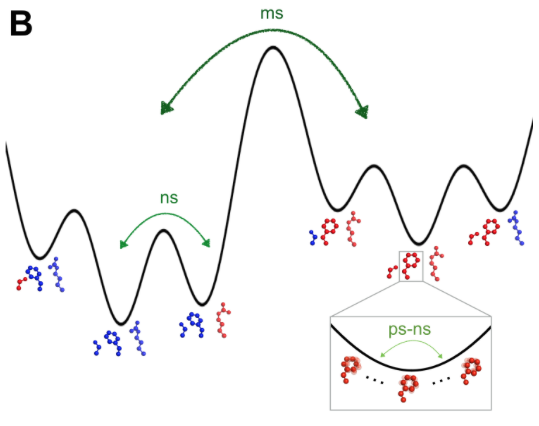
Determining the interconverting conformations of dynamic proteins in atomic detail is a major challenge for structural biology. Conformational heterogeneity in the active site of the dynamic enzyme cyclophilin A (CypA) has been previously linked to its catalytic function. Here we compare the conformational ensembles of CypA by fixed-target X-ray free electron laser (XFEL) crystallography and multitemperature synchrotron crystallography.
Mapping the Conformational Landscape of a Dynamic Enzyme by XFEL and Multitemperature Crystallography
Determining the interconverting conformations of dynamic proteins in atomic detail is a major challenge for structural biology. Conformational heterogeneity in the active site of the dynamic enzyme cyclophilin A (CypA) has been previously linked to its catalytic function. Here we compare the conformational ensembles of CypA by fixed-target X-ray free electron laser (XFEL) crystallography and multitemperature synchrotron crystallography. The “diffraction-before-destruction” nature of XFEL experiments provides a radiation-damage-free view of the functionally important alternative conformations of CypA. We monitored the temperature dependences of these alternative conformations with eight synchrotron datasets spanning 100-310 K. Multiconformer models show that many alternative conformations in CypA are populated above, but not below, the glass transition temperature (~200 K) and reveal abrupt changes in protein flexibility that provide all-atom insight into conformational coupling. Together, our XFEL data and multitemperature analyses motivate a new generation of time-resolved experiments to structurally characterize the dynamic underpinnings of protein function.