- Details
- Thursday, 16 February 2017

Membrane protein perform a variety of functions vital to the survival of an organism. As a result understanding how they work and why they malfunction has been an interest of researchers.
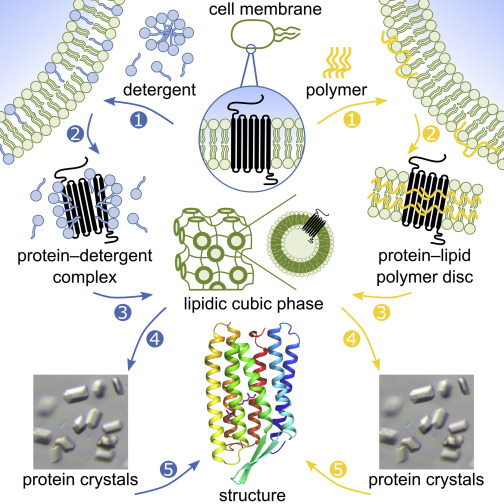
However determination of membrane protein structures has remained a challenge in large part due to the difficulty in establishing experimental conditions where the correct conformation of the protein in isolation from its native environment is preserved. This is partly due to the fact that detergents are often used to separate the protein from their lipid membrane encasing. Detergents often also strip away fat molecules crucial to stabilizing the protein. Researchers at the University of Toronto have discovered a new method for stabilizing membrane protein using a polymer originally developed for the auto industry.
Abstract
For some membrane proteins, detergent-mediated solubilization compromises protein stability and functionality, often impairing biophysical and structural analyses. Hence, membrane-protein structure determination is a continuing bottleneck in the field of protein crystallography. Here, as an alternative to approaches mediated by conventional detergents, we report the crystallogenesis of a recombinantly produced membrane protein that never left a lipid bilayer environment. We used styrene–maleic acid (SMA) copolymers to solubilize lipid-embedded proteins into SMA nanodiscs, purified these discs by affinity and size-exclusion chromatography, and transferred proteins into the lipidic cubic phase (LCP) for in meso crystallization. The 2.0-Å structure of an α-helical seven-transmembrane microbial rhodopsin thus obtained is of high quality and virtually identical to the 2.2-Å structure obtained from traditional detergent-based purification and subsequent LCP crystallization.
Introduction
Biological membranes contain a great variety of membrane proteins, which fulfill vital functions as receptors, signal transducers, channels, transporters, motors, and anchors (Fiedler et al., 2010). High-resolution structures of membrane proteins are still rare but are of paramount importance for understanding protein function on a molecular level with significant implications for biomedical and pharmaceutical applications (Bill et al., 2011). For in vitro studies such as biophysical analyses and structure determination, membrane proteins are usually purified with the aid of detergents. However, micelles formed by conventional detergents are poor membrane mimetics (Helenius and Simons, 1975), and membrane proteins in micelles do not experience the same lateral pressure profile as in a lipid bilayer (Van den Brink-van der Laan et al., 2004), which in many cases is important to maintain the native fold and function of the protein. Moreover, detergents tend to remove annular lipids, which are often essential for membrane-protein function (Paila et al., 2009). Accordingly, many membrane proteins are inactive in detergent micelles and require a membrane environment in order to be functional. Moreover, structural heterogeneity and conformational instability of detergent-solubilized membrane proteins can complicate quantitative as well as structural analyses (Privé, 2007)... Read full journal article here
