Popular Articles
- Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature
- NSF awards $22.5 million to capture biology at the atomic level using X-ray lasers
- BioXFEL Research Support Call for Proposals
- Frankuchen Award : ACA 2019
- UWM researchers create first 3D movie of virus in action
Archived Articles
- Details
- Thursday, 18 August 2016
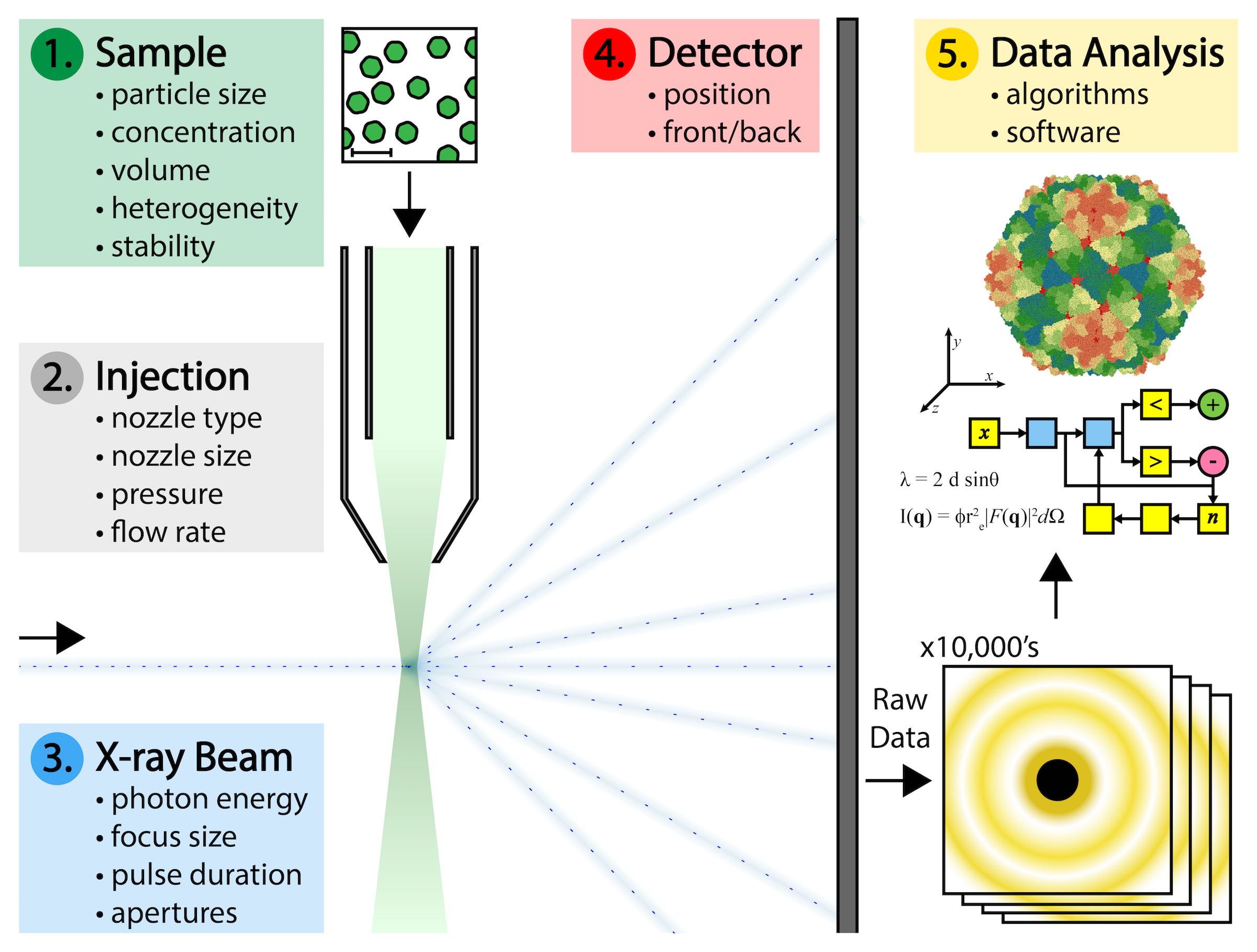 X-ray crystallography is a technique that can be used by scientists to map the 3-dimensional physical space of proteins and other biological molecules to an atomic level of detail. Over the last four decades or so, this technique has been used to map over 100,000 protein structures that can be found in the publicly accessible Protein Data Bank, with more being added every day.
X-ray crystallography is a technique that can be used by scientists to map the 3-dimensional physical space of proteins and other biological molecules to an atomic level of detail. Over the last four decades or so, this technique has been used to map over 100,000 protein structures that can be found in the publicly accessible Protein Data Bank, with more being added every day.
But the reach of X-ray crystallography is limited by its very nature because not all proteins are inclined to form crystals, and those that do are not guaranteed to behave in way that produces meaningful data. This has caused some scientists to investigate the prospect of skipping crystallography and measuring X-ray diffraction directly from the individual biological molecules, one particle at a time. Not only would this single particle approach open the door to a legion of important proteins, viruses, or other biological samples that have thus far evaded study, but it would also enable those samples to be investigated in real time under dynamic conditions that better reflects the true animated nature of biological molecules.
But significant technical challenges make this difficult to achieve. Biological samples must be produced at high concentrations and large volumes, and with as little variation as possible in the physical state of each individual particle. Specialized nozzle injection systems must be designed to deliver a continuous and adequate dispersion of particles to the X-ray beam without clogging or consuming the sample too quickly. Single particles do not diffract X-rays as intensely as crystals of those particles do. Single particles pass through the X-ray beam in random, unknown orientations that complicate the interpretation of the data they produce. And there are gigabytes of such data that result from a typical experiment that must be processed.
The answer to many of these challenges might be found in California’s Silicon Valley at a national laboratory called SLAC. Among other things, the campus at SLAC houses a 3-km long particle accelerator called an XFEL that began operations in 2009. The SLAC XFEL is capable of generating a continuous stream of X-ray pulses that are energetically a billion times more intense than what could be obtained from any X-ray instrument before it. Each such pulse of X-rays can be focused to a diameter of about 100 nanometers, which is about the size of a virus. The XFEL produces 120 of these X-ray pulses per second, and the duration of each pulse is measured in femtoseconds. For perspective, a femtosecond is a fraction of a second in the same way that a minute is a fraction of 1.9 billion years.
In the autumn of 2014, a group of researchers gathered for a meeting at SLAC to discuss ways to use the XFEL for imaging proteins and viruses. That meeting spawned a formal collaboration between over 100 scientists from 20 institutions in 8 different countries that became known as the Single Particle Imaging Initiative, or ‘SPI’. As stated in a roadmap penned by the leading participants, the shared goal of the collaboration was to “…solve the technical challenges related to single particle imaging with XFELs and to pave the way for single particle imaging with atomic resolution.”
Addressing those challenges required the coordination of experts from diverse fields such as microbiology, biophysics, fluid dynamics, mechanical engineering, optical physics, and computer programming. SPI researchers began by choosing a plant virus as the ideal sample to use throughout the ongoing series of experiments. Those experiments explored variations in the technical parameters related to preparing the sample, the mode of sample delivery, the X-ray beam characteristics, the beam detectors, and the interpretation and analysis of the resulting data.
Now after nearly two years of planning, communication and experimentation, the first report from the main researchers who participated in the SPI collaboration was released in the August 2016 volume of Nature’s Data Communications (linked here).
The group reported detection of X-ray diffraction at a level of 5.9 Å resolution, which is potentially sufficient for mapping a molecular level of detail. The report did not focus on fully interpreting the results, but rather made the raw data available in an open source fashion to any researchers who would like to become involved in the data analysis. Interpreting the data will require further experimentation with multiple computational algorithms that are capable of discerning, sorting and translating the raw data and weaving it together in 3-dimensional space to produce a single image that represents the cumulative measurements taken from thousands of individually diffracting particles.
The SPI collaboration will soon complete its course, leaving researchers free to apply the shared findings to their own collaborations or independent projects. These projects will continue at the SLAC XFEL, as well as at newer XFEL facilities that are now operational (or will soon be) in Japan, South Korea, Germany, and Switzerland.





