3. Accessing your data
Data Analysis
This resource belongs to the Data Analysis group.
Category
Published on
Abstract
Accessing your data at LCLS
The LCLS data analysis website is an excellent resource to help users do everything from setting up an account to analyzing their LCLS data with LCLS software. Our site is designed to help you with data analysis for serial femtosecond crystallography experiments at LCLS. Help with topics particularly for single particle and solution scattering experiments will come later.
Please refer to the LCLS Computing site and LCLS Data Analysis for updated information.
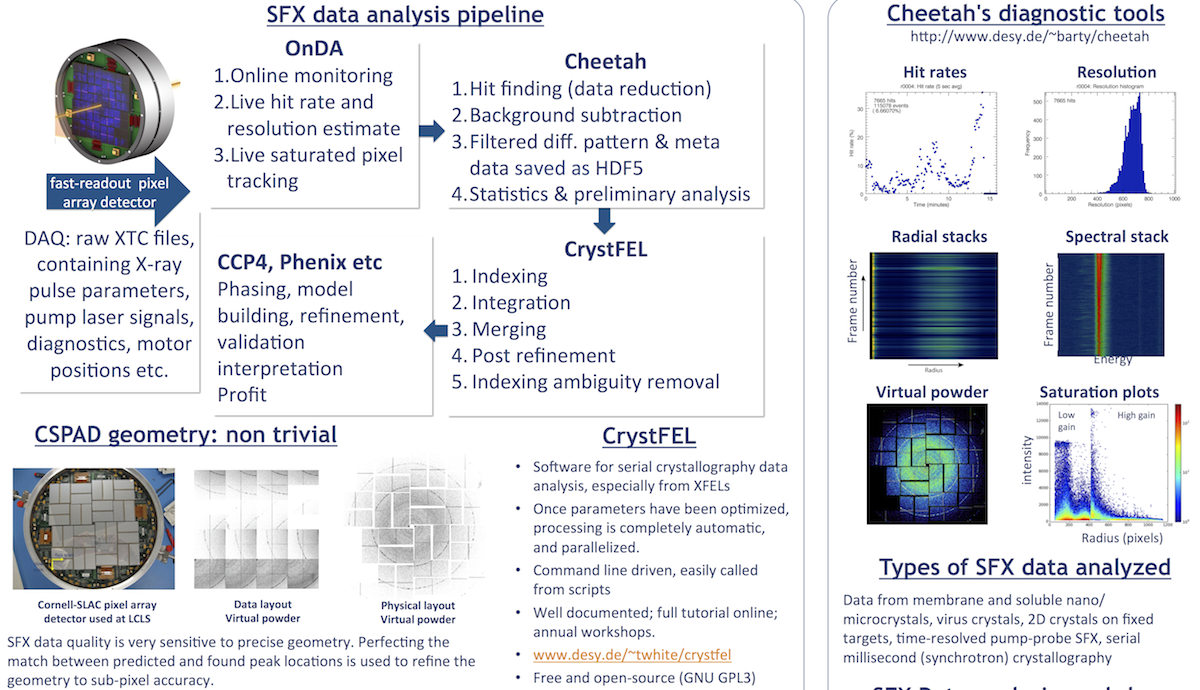
1. Learn about the basic data formats
Raw data from LCLS are written to the XTC format. This data is optimized to work with the data acquisition system, not for convenience of reading. Practically all experimental data are written to XTC files, and a set of such files are created for each "run" that is manually triggered by the experiment operator. LCLS provides tools if you wish to write your own code that interacts with XTC data, but for developed methods such as crystallography it is not common to do this.
Pre-processing programs like Cheetah can convert subsets of XTC data (just the "good" frames) to a format that is easier to work with in subsequent analysis such as structure factor integration. HDF5 is the usual and most recommended format. CXI is now a standard format and are just HDF5 files that follow a particular set of specifications intended for working with diffraction data.
Programs like CrystFEL read HDF5 and CXI data, and output various files. Cheetah also writes various "metadata" to text files for convenience of scripting.
2. Login on-site with an LCLS computer
If working on-site, you will be using one of the computers in the control room, or in the overflow / analysis room. Those computers do not have direct access to the data, so you must ssh into an interactive analysis node by typing:
$ ssh psana
Be sure to pick an analysis machine that is not being used by the beamline operators, the sample injector team, or for live data monitoring.
If your login is idle for a while, other users may log onto it and start running their analysis. You may check if someone is running jobs on it by typing:
$ top
If the node is too slow, try logging off and and logging back on again. The job handling system will attempt to connect you to the node that is least busy.
Note that the analysis nodes do not have access to the external internet, so you will not be able to transfer data to or from them. You can ssh into psexport to do that. See Data Management for more information.
3. Login remotely or from a personal laptop
The best way to access the LCLS machines remotely is via NoMachine as decribed here.
Using ssh with X forwarding is also possible, though slower.
$ ssh -X <username>@pslogin.slac.stanford.edu
To access and analyze data, log on to the interactive nodes:
$ ssh psana
this will put you on an interactive psana node, which has access to the data, while pslogin doesn't.
4. Set up an analysis environment
LCLS provides startup scripts that configure your unix environment for data analysis. These scripts need to be run once every time you log in to a different computer. However, this can be done automatically by modifying your startup file. To do this, first check which shell you are using by typing
$ echo $SHELL
If you use a bash shell, add the following to your startup file located at ~/.bashrc
test -f /reg/g/psdm/etc/ana_env.sh && . /reg/g/psdm/etc/ana_env.sh
For csh or tcsh, the following should be added to ~/.cshrc (paste this as a single line)
if ( -f /reg/g/psdm/etc/ana_env.csh ) source /reg/g/psdm/etc/ana_env.csh
There are additional configuration scripts for setting up Cheetah and CrystFEL at LCLS, which will be discussed in more detail later. For now, you may wish to setup Cheetah and CrystFEL by typing
$ source /reg/g/cfel/cheetah/setup.sh
$ source /reg/g/cfel/crystfel/crystfel-dev/setup-sh
at the command prompt or by including those lines in your ~/.bashrc file (if you are using the bash shell - you will find similar scripts for csh in the same locations).
5. Locate your data
If your experiment is conducted at CXI and your proposal number is LC49 and it was carried out in 2014, then your data will be in a directory named
/reg/d/psdm/cxi/cxic4914
The raw XTC files will be in the sub-directory
/reg/d/psdm/cxi/cxic4914/xtc
Your scratch space will be in
/reg/d/psdm/cxi/cxic4914/scratch
The scratch space is where you should write most of your temporary files generated during analysis (e.g. files generated by Cheetah). You can do whatever you want with your scratch space, but it is a good idea to follow conventions to help streamline analysis. Make a sub-directory for yourself here, preferably with your username. If you follow the subsequent setup steps in this tutorial, your Cheetah output will be in
/reg/d/psdm/cxi/cxic4914/scratch/<username>/cheetah/hdf5
The sub-directories associated with your experiment are different "storage classes" so LCLS can manage their data storage and backups. The table below is correct as of November 2016 - see the LCLS data retention policy for more details. XTC files will remain on disc for 4 months, and then move to tape for 10 years (they can later be restored to disk at about 1 TB/hr). The scratch space is "unlimited", but is not backed up and also has a lifetime of only 4 months. If you want your data to persist longer, you might want to keep your text-based scripts and configuration files in the results space, and copy your best diffraction data to the ftc directory. Symbolic links are helpful when setting up your directory structure.
|
Space |
Quota |
Backup |
Lifetime |
Comment |
|---|---|---|---|---|
|
xtc |
None |
Tape archive |
4 months |
Raw data |
|
usrdaq |
None |
Tape archive |
4 months |
Raw data from users' DAQ systems |
|
hdf5 |
None |
Tape archive |
4 months |
Data translated to HDF5 |
|
scratch |
None |
None |
4 months |
Temporary data (lifetime not guaranteed) |
|
results |
4TB |
Tape backup |
2 years |
Analysis results |
| calib | None | Tape backup | 2 years | Calibration data |
|
User home |
20GB |
Disk + tape |
Indefinite |
User code |
|
Tape archive |
- |
- |
10 years |
Raw data (xtc, hdf5, usrdaq) |
| Tape backup | - | - | Indefinite | User home, results and calib folder |
| Disk backup | - | - | Indefinite |
Accessible under ~/.zfs/ |
Note that the LCLS-generated hdf5 directory
/reg/d/psdm/cxi/cxic4914/hdf5
does not contain the output of programs like Cheetah. Only HDF5's created by the XTC-to-HDF5 translator created by LCLS are stored here. This conversion is rarely used in SFX experiments.
6. Submitting batch jobs
This can be done on site or remotely. During your LCLS experiment, you have priority access to LCLS computing resources, some of which have been set aside ONLY for current experiments.
Unless you are currently running an experiment, i.e. if you are accessing your data later or are participating in a workshop, please DO NOT SUBMIT JOBS TO QUEUES LABELLED WITH "PRIO", e.g. psnehprioq or psfehprioq. These are priority queues for current, running experiments only. There are plenty of other computing nodes available to you that will not interfere with current LCLS users.
For details on all batch nodes, check https://confluence.slac.stanford.edu/display/PCDS/Submitting+Batch+Jobs
To check if your jobs are running
$ bjobs
To see the status of all jobs, or all jobs on the queue "psanaq"
$ bjobs -u all
$ bjobs -u all | grep "psanaq"
Please check the LCLS Computing site and LCLS Data Analysis for updated information.
7. Other ways to inspect your data
Python is the most recommended way to write your own analysis code (because many are now switching to Python, and it is well-supported at LCLS). You can access LCLS data via the psana: python interface. Most crystallography users will not need to write their own code for working with XTC data.
If you need Matlab, look here. There are only two analysis nodes that support matlab.
If you need IDL, look here. There is currently only one license for IDL at SLAC.
References
PREVIOUS: 2. During your beamtime NEXT: 4. Detectors and geometry
SKIP TO: 5. Hit finding and data reduction with Cheetah
Back to front page: LCLS serial femtosecond crystallography data analysis instructions