Popular Articles
- Earliest molecular events of vision revealed
- Dynamics and Kinetics in Structural Biology
- Structural biology is solved -- now what?
- XFEL Pulses Demonstrate How Plants Perceive Light
- BioXFEL Postdoctoral Fellowship Award
Archived Articles
News
- Details
- Wednesday, 28 August 2024
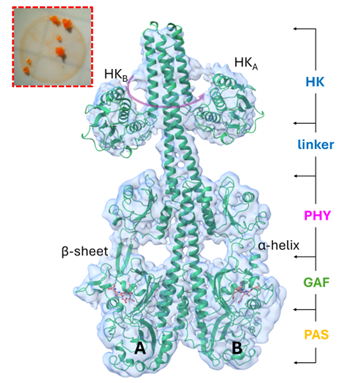
A group of BioXFEL researchers brought single particle cryoEM structure determination to BioXFEL. The research was spearheaded by Tek Malla, a BioXFEL graduate in the Schmidt lab.
- Details
- Tuesday, 21 November 2023

Understand the latest experimental tools in structured biology with this pioneering work by Keith Moffat and Eaton E. Lattman. Structural biology seeks to understand the chemical mechanisms and functions of biological molecules based on their atomic structures. Until recently, these structures have been studied only statically.
- Details
- Wednesday, 18 October 2023
"Visualizing the 3D Structures of Molecules Involved in Life's Processes" Workshop
Undergraduate students attending college in Puerto Rico are invited to participate in this two-day workshop.
The workshop will be available in the following locations from 8am - 4pm each day:
- February 5th - 6th in Mayaguez
- February 7th - 8th in San Juan
Workshop Components:
-
Focused on structural biology at X-ray sources
-
Lectures and practicals from experts in the field
-
Real synchrotron data collection and data processing opportunities
General Workshop Topics:
-
Introduction to Structural Biology
-
Crystallographic approaches
-
X-ray imaging at synchrotrons and XFELs)
-
Complementary techniques
- Details
- Wednesday, 18 October 2023
The 2024 BioXFEL Spring Symposium will be at Arizona State University from March 6 – 7, 2024. The symposium will consist of scientist and student presentations, a poster session, and networking/recruitment opportunities.
For registration payment and information, please click here.

- Details
- Friday, 08 September 2023
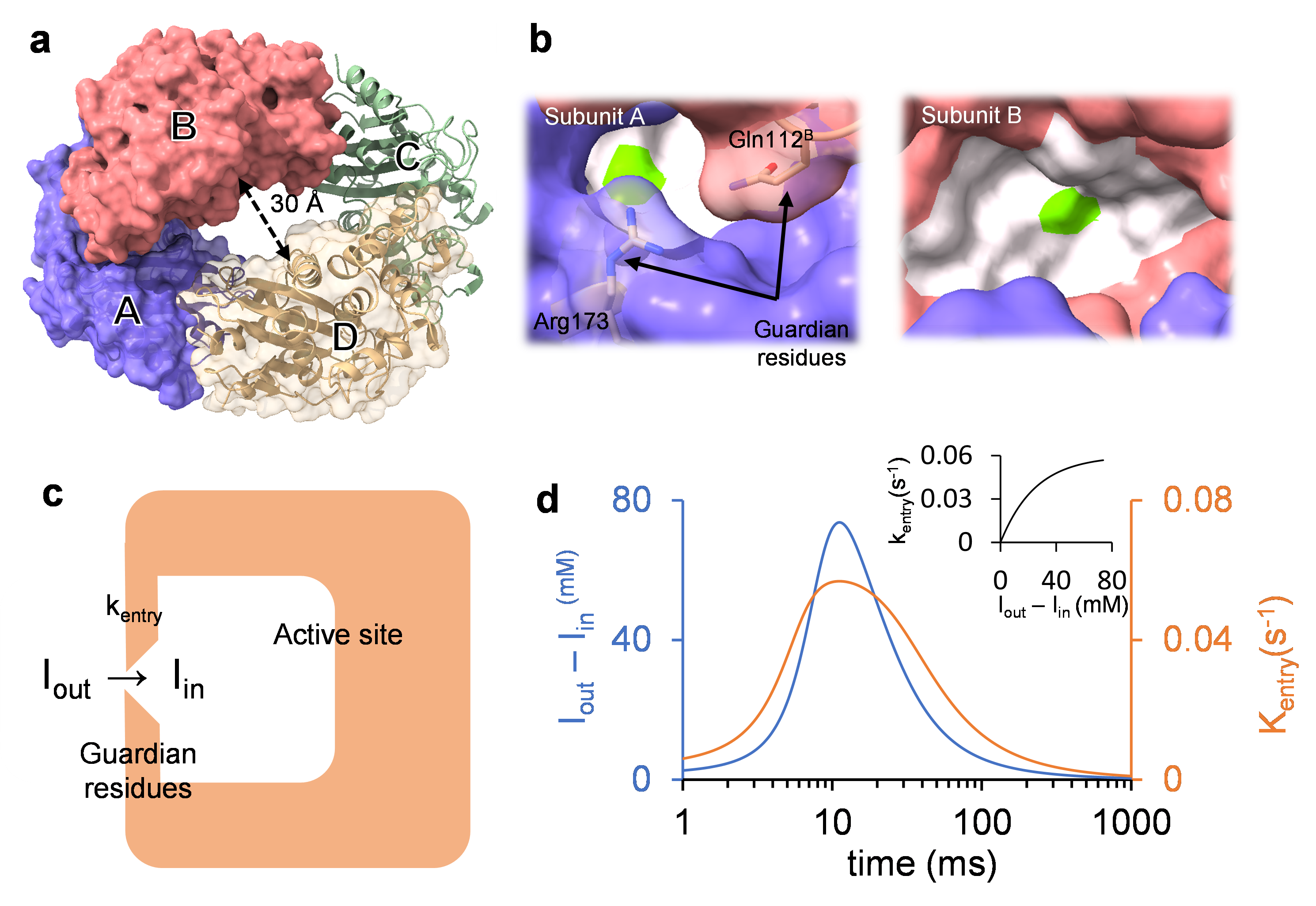
In a paper just published in Nature Communications, a team of BioXFEL researchers and collaborators used mix-and-inject serial crystallography (MISC) to analyze the reaction of the β-lactamase BlaC from tuberculosis bacteria with the suicide inhibitor sulbactam (SUB).
- Details
- Thursday, 23 March 2023
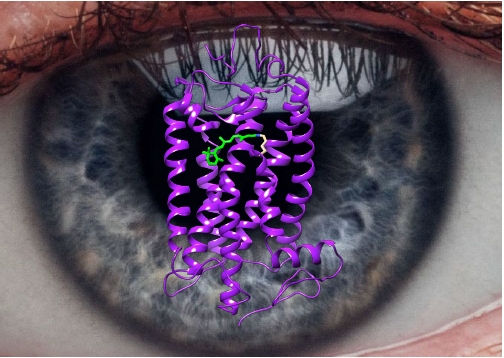
A large international team lead by Valerie Paneels and Gebhard Schertler revealed the molecular mechanism of light receptor activation in the eyes of vertebrae animals by time-resolved serial femtosecond crystallography (TR-SFX). The article appeared in Nature online on March 22 2023. BioXFEL researchers Schmidt and Stojković were invited to write a News and Views article summarizing the general findings focused on the light-activated bovine rhodopsin.
The News and Views article is available here: https://www.nature.com/articles/d41586-023-00504-4.
Figure
Caption: The structure of the eye-pigment rhodopsin in the resting state. The retinal is shown in green in the 11-cis configuration. The retinal is bound via a Schiff-base to a lysine sidechain. BioXFEL researchers Schmidt and Stojković wrote a News and Views article in Nature summarizing newest results on the activation of rhodopsin by light.






