Introduction to serial femtosecond crystallography data analysis
Data Analysis
This resource belongs to the Data Analysis group.
Category
Published on
Abstract
Click or use the left and right arrow keys to advance forward and backward in this embedded presentation (downloadable PDF available in the Supporting Docs tab above)
Serial femtosecond crystallography with X-ray free electron lasers
X-ray free electron lasers (XFELs) have enabled biomolecular nano- and micro-crystallography at ambient temperatures by using extremely brief X-ray pulses (each only a few tens of femtoseconds) to outrun radiation damage, which is an inherent problem in bio-imaging techniques. The X-ray pulses used in this "diffract and destroy" mode are so intense that only a single shapshot diffraction pattern is obtained from a crystal before it is destroyed (after a useful image has been acquired). The crystal supply must thus be replenished continuously, in a serial manner, and the detector read out after each shot. This methodology, serial femtosecond crystallography (SFX), has yielded several major and unique advances in structural biology previously unattainable with conventional technologies, including the potential for sub-picosecond time-resolved crystallographic studies, probing cyclic or even non-cyclic reactions.
In SFX, nano/microcrystals are delivered to the pulsed X-ray beam by either a micron-thick liquid jet (most commonly), a recently invented lipidic cubic phase (LCP) jet, electrospray, or by raster scanning fixed target supports. At LCLS, the X-ray pulses arrive at 120 Hz (with much higher rates planned for future XFELs), resulting in huge data sets that need to be efficiently filtered into crystal hits and empty frames, with various detector corrections applied, leading to collections of diffraction patterns totally 10's to 100's of terabytes.
Each diffraction pattern is then indexed and integrated independently, before the data set is merged into a reflection list to be used for phasing. Thousands of diffraction patterns, and hence crystals, are needed for a full data set. For time-resolved studies, thousands of randomly oriented patterns are needed for each time point. A single experiment can result in over 100 terabytes of raw data. These high data collection rates and large data sets have necessitated the development of novel high-throughput, parallelizable data analysis software for "live" feedback during an SFX experiment, as well as for further processing of the clean diffraction patterns. Since the diffraction patterns consist almost entirely of partial reflections, complicated post-refinement/scaling procedures are necessary to significantly reduce the amount of data required to obtain accurate structure factors. A number of post refinement methods are being developed by the SFX community.
With the construction of more than a dozen new XFELs currently under way, and the multiple recent demonstrations of SFX at synchrotrons, the potential user base is growing significantly. The development and appropriate use of new software to tackle the unique problems of SFX data analysis is vital to making SFX practicable.
Recent reviews for serial crystallography
- Spence, J. C. H. X-ray lasers and serial crystallography. IUCrJ 2, 305–306, (2015). . http://journals.iucr.org/m/issues/2015/03/00/me0576/index.html
- Schlichting, I. Serial Femtosecond crystallography: the first five years. IUCrJ. 2, 246-255. http://journals.iucr.org/m/issues/2015/02/00/it5004/
The first demonstration of serial femtosecond protein nanocrystallography
- Chapman HN. et al. 2011 Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77. (doi:10. 1038/nature09750)
Theoretical aspects of structure factor analysis in serial femtosecond crystallography
- Kirian RA et al. 2010 Femtosecond X-ray protein nanocrystallography: data analysis methods. Opt. Express. 18, 5713–5723. (doi:10.1364/OE.18. 005713)
- Kirian RA et al. 2011 Structure-factor analysis of femtosecond micro-diffraction patterns from protein nanocrystals. Acta Crystallogr. A 67, 131 – 140. (doi:10.1107/S0108767310050981)
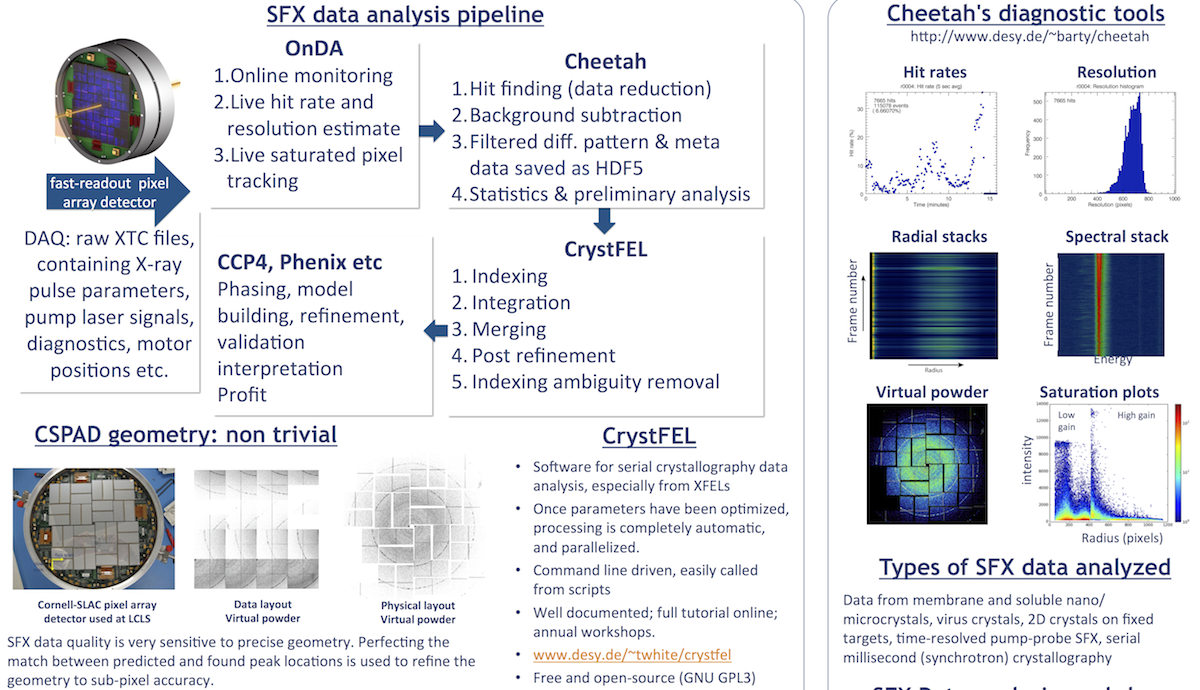
Data reduction, hit finding, data quality estimates
OnDA includes an online, live hit rate and saturation rate monitor (during data collection), as well as an interactive peak finding parameter tweaker for use with Cheetah. OnDA was written by Valerio Mariani at the Center for Free Electron Laser Science at DESY. Details on how to set up OnDA are here:
Cheetah is set of programs for processing serial diffraction data data from at free electron laser sources, and which enable taking home only the data with meaningful content. Detailed instructions on how to get the Cheetah GUI running in your LCLS experiment are available on the Cheetah website. Cheetah development is led by Anton Barty, at the Center for Free Electron Laser Science at DESY. The main reference for Cheetah is
- A. Barty, R. A. Kirian, F. R. N. C. Maia, M. Hantke, C. H. Yoon, T. A. White, and H. N. Chapman, "Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data," J Appl Crystallogr, vol. 47, pp. 1118–1131 (2014). doi:10.1107/S1600576714007626 - Download PDF
Indexing, merging and post-refinement
CrystFEL is a suite of programs for processing diffraction data acquired "serially" in a "snapshot" manner, such as when using the technique of Serial Femtosecond Crystallography (SFX) with a free-electron laser source. CrystFEL development is led by Thomas White, at the Center for Free Electron Laser Science at DESY. The main references for CrystFEL are
- White TA, Kirian RA, Martin AV, Aquila A, Nass K, Barty A, Chapman HN. 2012 CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341. (doi:10.1107/S00218898 12002312)
- White TA, Barty A, Stellato F, Holton JM, Kirian RA, Zatsepin NA, Chapman HN. 2013 Crystallographic data processing for free electron laser sources. Acta Crystallogr. D 69, 1231 – 1240. (doi:10.1107/ S0907444913013620) download PDF
- T. A. White. "Post-refinement method for snapshot serial crystallography". Phil. Trans. Roy. Soc. B 369 (2014) 20130330. doi:10.1098/rstb.2013.0330 (open access - download PDF).
CrystFEL has excellent up-to-date documentation, detailed man (manual) pages, a thorough tutorial including real SFX data from LCLS, a "best practices" page and FAQ on its website. The CrystFEL tutorial is here: http://www.desy.de/~twhite/crystfel/tutorial.html
Data analysis scripts
A repository of useful scripts for the analysis of XFEL data is available here: www.bioxfel.org/science/resources/scripts