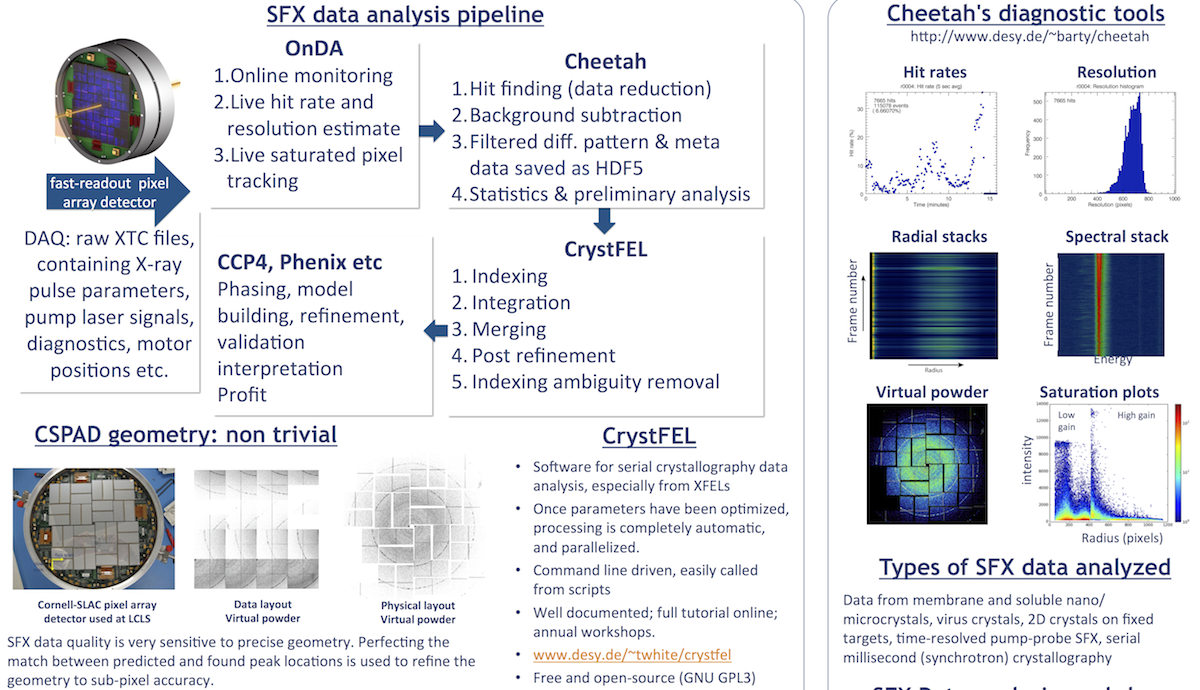
1. Before your beamtime
Data Analysis
This resource belongs to the Data Analysis group.
Category
Published on
Abstract
Before your beamtime
1. Get an account in the LCLS system.
You need a SLAC user portal account (you must be listed as a collaborator on a proposal to get one), and you also need a unix account. Follow the instructions here: https://confluence.slac.stanford.edu/display/PCDS/Accounts
Make sure your SLAC Cyber Security Training is up to date, even if you will not be on site (e.g. accessing data remotely and for workshops).
Once your account is set up, check that you actually have access to the experiment. A good link to remember is https://pswww.slac.stanford.edu/ ; from there, click on the Data Manager link followed by the link to your experiment number. Importantly, you will find a link to the electronic logbook (eLog) for the experiment. If you do not see a link to your experiment, ask the PI to add you to the experiment via the user management tool.
2. Set up a spreadsheet for the experiment.
We highly recommend setting up a log (e.g. Google Drive sheets) for keeping track of runs, samples, and all sorts of experimental metadata. Here's an example of the metadata you may find worth keeping. Even for protein screening beamtimes, a single spreadsheet is very handy for data analysts later, rather than searching through the eLog for comments on each run.
Download a template spreadsheet here.
| Run # | Time (start) | Duration | Sample | sample volume loaded | Comments | Trans-mission % | frames recorded | hits | hit rate (%) | Totals | resolution estimate [Å] | indexed | who's indexing | Rep. rate (Hz) | Photon Energy (keV) | Wavelength [A] | detector encoder position | Detector position (mm) | Pulse length (fs) ~ | Pulse energy (mJ) | Nozzle ID | Nozzle diameter | Distance between shots (um) | LCP jet speed (mm/s) | LCP Flow rate (nl/min) | Gas pressure (psi) | Pressure on liquid (psi) | Flow rate (ul/min) | Filter (um) | Priority |
| 1 | 9:00 AM | 0:00:30 | DARK | - | - | - | - | - | - | - | - | - | - | 120 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 2 | 11:30 AM | 0:10:00 | calibration sample, e.g. lysozyme xtals | 33 | 2 | 120 | 9.5 | 1.3 | -490 | 80 | 45 | 1 | ||||||||||||||||||
| 3 | 11:43 AM | 0:12:39 | name of sample | 15 µL | 5 | 91080 | 9108 | 10 | 4 | 7302 | nadia | 120 | 9.5 | 1.3 | -490 | 80 | 45 | 2 | 50 µm | 1 |
3. Learn at least the basic unix commands
You will some basic unix commands to access your data and be able to set up and run Cheetah, OnDA, and CrystFEL.
New to unix? Here are some resources:
1. James Holton put together a good list of basics unix commands (and explanations) that everyone should know. Learn them.
2. Man pages (manual pages) are really helpful for linux commands. Here is a full list of them: http://man7.org/linux/man-pages/dir_all_alphabetic.html
Many Linux programs have man pages easily accessible via "man <program name>".
3. There are many other good introductions to unix commands online, e.g. http://faculty.tru.ca/nmora/Frequently%20used%20UNIX%20commands.pdf
4. Discuss what parameters you will need to save in the raw data stream.
It is very important that the data analysts, in particular, have a discussion long before the beam time that actually explains what parameters they need to work with the data, before they go to the beamline scientist to make sure those data are selected.
Many users have come with the intention of saving much less data than is advisable.
References
NEXT: 2. During the beamtime.
Back to front page: LCLS serial femtosecond crystallography data analysis instructions